Multimodal Causal Learning and Reasoning for Secondary Health Conditions in SCI Individuals
Conditions such as pulmonary infections, pressure injuries (PI), urinary tract infections, and cardiovascular disease are highly prevalent among the SCI population, imposing a huge risk on the person’s health. However, there is a lack of direct measurement methods and sufficient data for modelling and defining biomarkers of complex interactions among co-morbidities. Nonetheless, continuous monitoring has the potential to enable observing multiple complex interactions between behavioural and physiological data.
In this project, we aim to construct a causal representation that can abstract the physiological mechanisms from multimodal data, which could help us understand the dynamics and development of comorbidities. Causal graphical models can serve as a rigorous statistical framework to represent the body processes in an organized and compact manner, identifying less spurious relationships and increasing robustness against domain shifts in multiple populations.
As a rigorous statistical modelling framework, graphical models can represent the relations of variables in an explicit and explainable manner, which become especially needed in health care and biomedical research where a traceable representation of knowledge is expected. Based on the structural representation, we are developing an explainable and powerful detection system to fuse clinical and sensor data for personalized risk identification of SCI comorbidities.
Under this framework, nodes in the graph represent the variables depicting the body conditions such as onsets of PI/AD and high-level features (a proxy for variables) extracted from biomarkers. Each node is random in nature and conditionally dependent on a certain node(s). Based on the topological structure, information can be passed from leaf nodes to nodes of interest to deduce the likelihood (probability of taking certain values) of event occurrence. The graph is expected to be multi-modal, which accommodates both static variables (demographic characteristics of patients) and dynamic features (extracted signals from biomarker sensors).
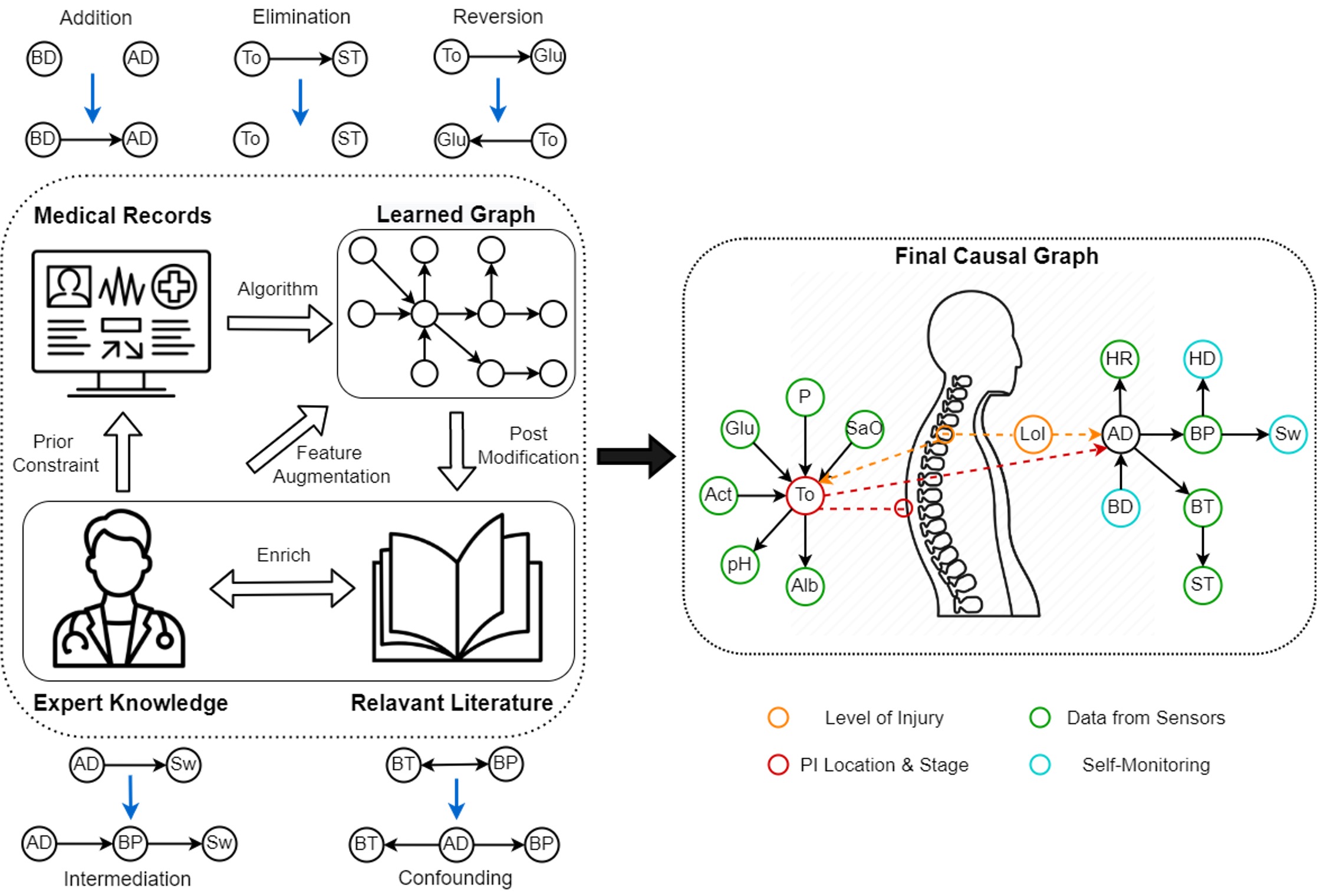
Causal discovery (discovering the causal relations from data) has always been a challenging problem due to the huge discrepancy between idealistic assumptions and realistic complexities, calling for methodological improvement and innovation. Therefore, we propose to learn causal relations through a human-in-the-loop manner (as depicted above) combining both data-driven approaches and iterative incorporation of expert knowledge. Based on the discovered causal structures we could develop models of the risk of disease onset to be inferred from the learned causal graph and novel dynamic data inputs.
In this project, we propose to constrain the data-driven causal discovery with auxiliary expert knowledge in an iterative way. Our focus would be to improve the current knowledge discovery & representation by combining different information sources and to develop a pipeline of continual causal learning and inference for hospitals/institutes in different diseases.
Pressure Injuries Causal Discovery
Pressure Injuries are tissue injuries, sores or ulcers that happen on areas of the skin that are under pressure usually over bone prominences. Among the SCI population, this is a highly prevalent co-morbidity because of the long time spent on a wheelchair or a bed without pressure release. Due to the loss of sensation, persons with SCI do not perceive the corresponding warning signs such as pain thus their PI could escalate to much more severe stages without any intervention.
Despite much research on PI, they are a complex process still unclear. The combination of shear and friction, the time and frequency of applied pressure, and the soft tissue’s characteristics and oxygenation level are significant factors. Nonetheless, an individual’s medical history, etiology of SCI and time since injury, muscle build, posture, nutritional status, and many other behavioural factors are as well significantly contributing confounders that must be accounted for. Our objective is to identify the causal relations among those interested factors towards the onsets and development of PI in different locations, through clinical data gathered from different patient groups divided according to their purposes of hospital stay. We would test both constraint-based methods and score-based methods for causal discovery and development and an interactive system for doctors to input their expert knowledge and edit learned graphs from data.
The use of the high-level causal graph can enable explicit knowledge representation and a structural foundation for building a more fine-grained dynamic representation of PI evolution. This output can be incorporated with a broad range of bio-signals (e.g., HR, RR, ECG, Temperature, Oxygenation, Movement, etc.) for continuous, unobtrusive, and reliable data capturing from individuals in both clinical and outpatient settings during their daily life, herewith, opening the path towards a personalized risk model for PI development at specific body areas with both high accuracy and strong interpretability.
Founded by the ETH-SPS Digital Transformation in Personalized Health Care for SCI Project
Working Group
- Yanke Li
- Yahang Qi
- Ayoube Ighissou
- Jonathan Koch
External Collaborators
- external page KD Dr. Med. PhD Anke Scheel-Sailer (SPZ - Inpatient Unit)
- Dr. Med Benjamin Wiludda (SPZ)